CARD Overview
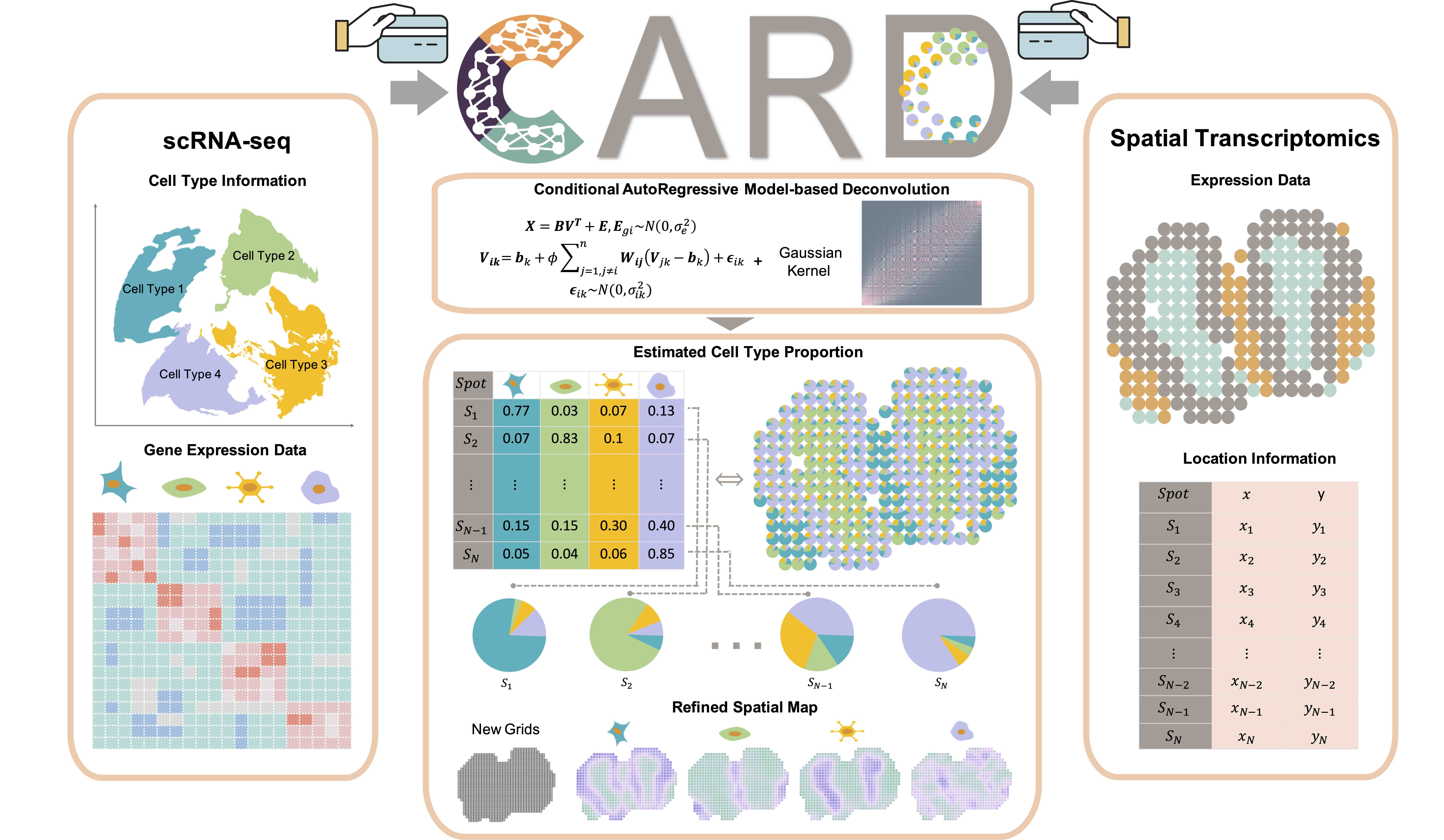 CARD is a R package developed to perform spatially informed cell type deconvolution for Spatial Transcriptomics. Briefly, CARD is a reference-based deconvolution method that estimates cell type composition in spatial transcriptomics based on cell type specific expression information obtained from a reference scRNA-seq data. A key feature of CARD is its ability to accommodate spatial correlation in the cell type composition across tissue locations, enabling accurate and spatially informed cell type deconvolution as well as refined spatial map construction. CARD relies on an efficient optimization algorithm for constrained maximum likelihood estimation and is scalable to spatial transcriptomics with tens of thousands of spatial locations and tens of thousands of genes. CARD is implemented as an open-source R package, freely available at www.xzlab.org/software.html.
CARD is a R package developed to perform spatially informed cell type deconvolution for Spatial Transcriptomics. Briefly, CARD is a reference-based deconvolution method that estimates cell type composition in spatial transcriptomics based on cell type specific expression information obtained from a reference scRNA-seq data. A key feature of CARD is its ability to accommodate spatial correlation in the cell type composition across tissue locations, enabling accurate and spatially informed cell type deconvolution as well as refined spatial map construction. CARD relies on an efficient optimization algorithm for constrained maximum likelihood estimation and is scalable to spatial transcriptomics with tens of thousands of spatial locations and tens of thousands of genes. CARD is implemented as an open-source R package, freely available at www.xzlab.org/software.html.